Retrieve (as an rspec object) or plot pavo's in-built spectral sensitivity data.
Usage
sensdata(
visual = c("none", "all", "avg.uv", "avg.v", "bluetit", "ctenophorus", "star", "pfowl",
"apis", "canis", "cie2", "cie10", "musca", "drosophila", "habronattus",
"rhinecanthus"),
achromatic = c("none", "all", "bt.dc", "ch.dc", "st.dc", "md.r1", "dm.r1", "ra.dc",
"cf.r"),
illum = c("none", "all", "bluesky", "D65", "forestshade"),
trans = c("none", "all", "bluetit", "blackbird"),
bkg = c("none", "all", "green"),
plot = FALSE,
...
)Arguments
- visual
visual systems. Options are:
"none": no visual sensitivity data."all": all visual sensitivity data."apis": Honeybee Apis mellifera visual system."avg.uv": average avian UV system."avg.v": average avian V system."bluetit": Blue tit Cyanistes caeruleus visual system."canis": Canid Canis familiaris visual system."cie2": 2-degree colour matching functions for CIE models of human colour vision. Functions are linear transformations of the 2-degree cone fundamentals of Stockman & Sharpe (2000), as ratified by the CIE (2006)."cie10": 10-degree colour matching functions for CIE models of human colour vision. Functions are linear transformations of the 10-degree cone fundamentals of Stockman & Sharpe (2000), as ratified by the CIE (2006)."ctenophorus": Ornate dragon lizard Ctenophorus ornatus."musca": Housefly Musca domestica visual system.'drosophila': Vinegar fly Drosophila melanogaster (Sharkey et al. 2020)."pfowl": Peafowl Pavo cristatus visual system."star": Starling Sturnus vulgaris visual system."habronattus": Jumping spider Habronattus pyrrithrix."rhinecanthus": Triggerfish Rhinecanthus aculeatus.
- achromatic
the sensitivity data used to calculate luminance (achromatic) receptor stimulation. Options are:
"none": no achromatic sensitivity data."all": all achromatic sensitivity data."bt.dc": Blue tit Cyanistes caeruleus double cone."ch.dc": Chicken Gallus gallus double cone."st.dc": Starling Sturnus vulgaris double cone."cf.r": Canid Canis familiaris rod"md.r1": Housefly Musca domestica R1-6 photoreceptor.'dm.r1': Vinegar fly Drosophila melanogaster R1-6 photoreceptor."ra.dc": Triggerfish Rhinecanthus aculeatus double cone.
- illum
illuminants. Options are:
"none": no illuminant data."all": all background spectral data."bluesky"open blue sky."D65": standard daylight."forestshade"forest shade.
- trans
Ocular transmission data. Options are:
"none": no transmission data."all": all transmission data."bluetit": blue tit Cyanistes caeruleus ocular transmission (from Hart et al. 2000)."blackbird": blackbird Turdus merula ocular transmission (from Hart et al. 2000).
- bkg
background spectra. Options are:
"none": no background spectral data."all": all background spectral data."green": green foliage.
- plot
should the spectral data be plotted, or returned instead (defaults to
FALSE)?- ...
additional graphical options passed to
plot.rspec()whenplot = TRUE.
Value
An object of class rspec (when plot = FALSE), containing
a wavelength column "wl" and spectral data binned at 1 nm intervals from 300-700 nm.
References
Sharkey, C. R., Blanco, J., Leibowitz, M. M., Pinto-Benito, D., & Wardill, T. J. (2020). The spectral sensitivity of Drosophila photoreceptors. Scientific reports, 10(1), 1-13.
Examples
# Plot the honeybee's receptors
sensdata(visual = "apis", ylab = "Absorbance", plot = TRUE)
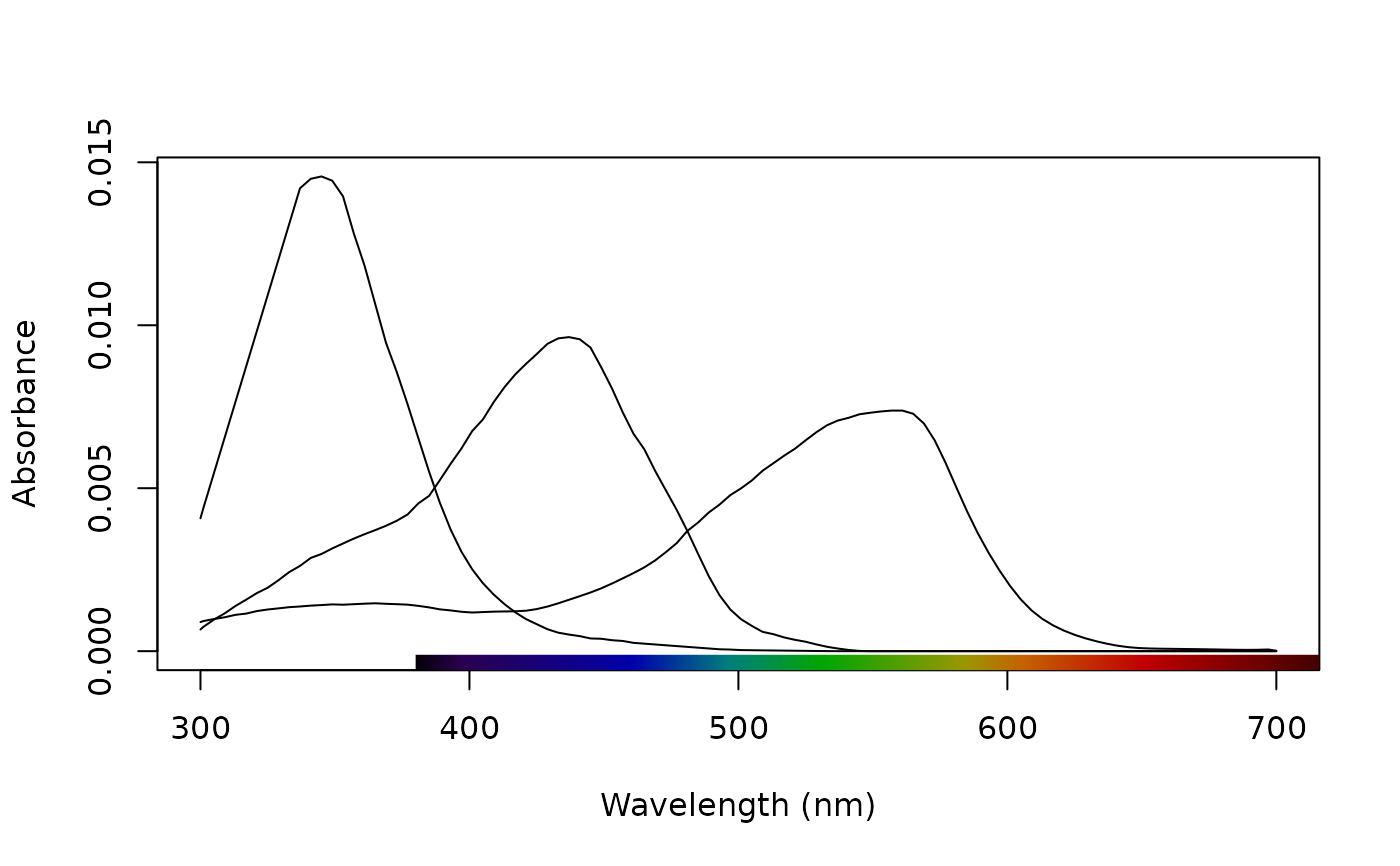 # Plot the average UV vs V avian receptors
sensdata(visual = c("avg.v", "avg.uv"), ylab = "Absorbance", plot = TRUE)
# Plot the average UV vs V avian receptors
sensdata(visual = c("avg.v", "avg.uv"), ylab = "Absorbance", plot = TRUE)
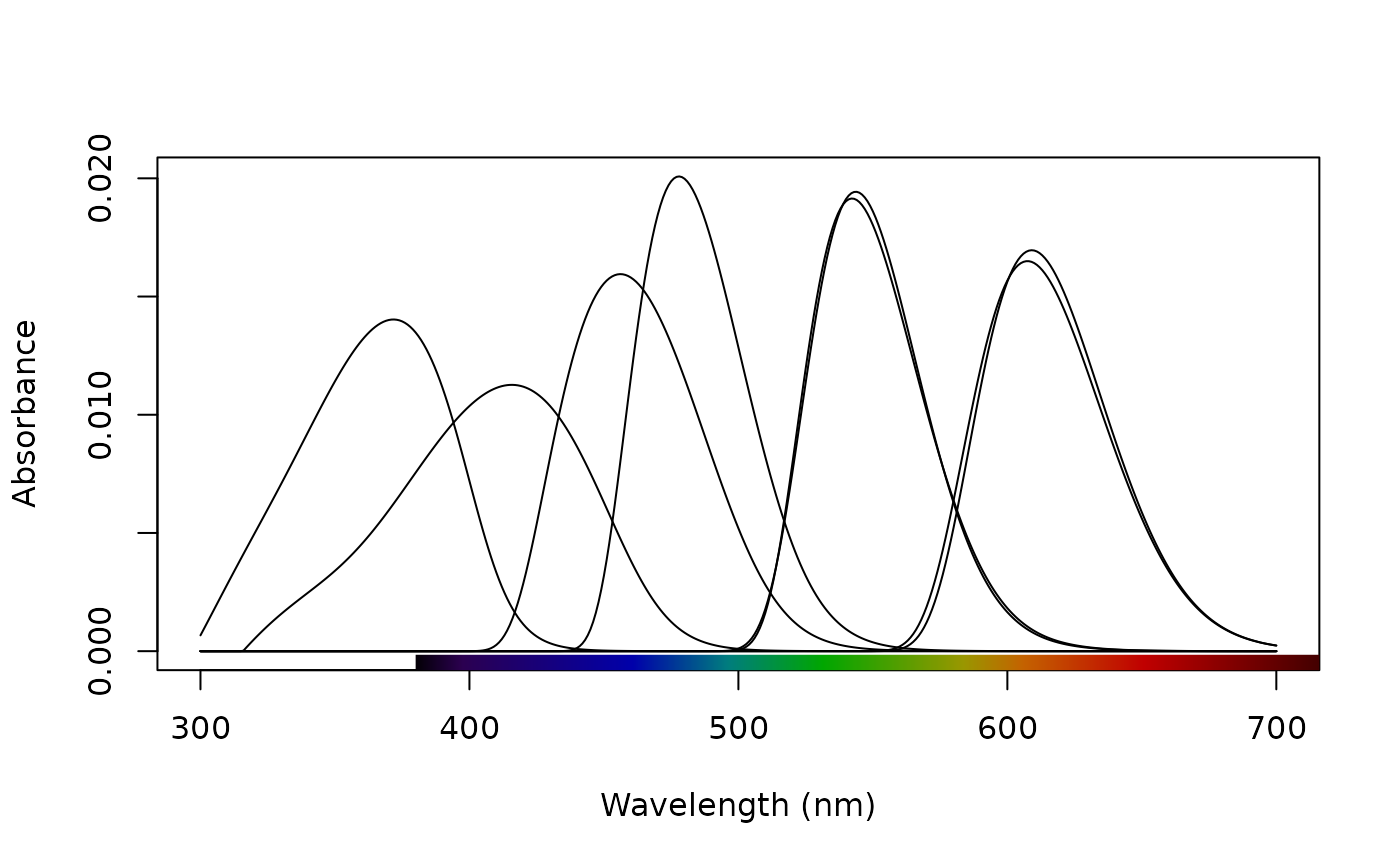 # Retrieve the CIE colour matching functions as an rspec object
ciedat <- sensdata(visual = c("cie2", "cie10"))
# Retrieve the CIE colour matching functions as an rspec object
ciedat <- sensdata(visual = c("cie2", "cie10"))